# Load library
library(tidyverse)How to perform efficient simulations in Tidyverse
Use purrr::map() for element-wise operations
R Programming
Tidyverse
Simulations
General steps
- Generate multiple datasets: Simulate \(N\) datasets based on a defined data-generating process.
- Apply a statistical method: For each dataset, apply the same statistical method (e.g., linear regression, hypothesis testing).
- Summarize the results: Aggregate and summarize performance metrics (e.g., bias, MSE, coverage).
Example: linear regression
Consider a simulation study for linear regression.
Define the data-generating process
Simulate a dataset for linear regression:
generate_data <- function(n = 100, beta0 = 1, beta1 = 2, sigma = 1) {
tibble(
x = rnorm(n),
y = beta0 + beta1 * x + rnorm(n, sd = sigma)
)
}Define the statistical method
Fit a linear model and extract the coefficient estimates:
fit_model <- function(data) {
model <- lm(y ~ x, data = data)
tibble(
beta0_hat = coef(model)[1],
beta1_hat = coef(model)[2]
)
}Perform simulation
Generate \(N = 1000\) datasets, each with an id (sim_id) and stored in a list-valued column data.
set.seed(123)
# Number of simulations
N <- 1000
# Simulate data
simulated_data <- tibble(
sim_id = 1:N
) %>%
mutate(
data = map(sim_id, ~ generate_data(n = 100, beta0 = 1, beta1 = 2, sigma = 1)) # Generate datasets
)
# Take a look
simulated_data
#> # A tibble: 1,000 × 2
#> sim_id data
#> <int> <list>
#> 1 1 <tibble [100 × 2]>
#> 2 2 <tibble [100 × 2]>
#> 3 3 <tibble [100 × 2]>
#> 4 4 <tibble [100 × 2]>
#> 5 5 <tibble [100 × 2]>
#> 6 6 <tibble [100 × 2]>
#> 7 7 <tibble [100 × 2]>
#> 8 8 <tibble [100 × 2]>
#> 9 9 <tibble [100 × 2]>
#> 10 10 <tibble [100 × 2]>
#> # ℹ 990 more rows
#> # ℹ Use `print(n = ...)` to see more rowsUse purrr::map() to fit a linear model to each entry of data and store the results in a list-valued column results. Then unnest to combine all results into a single table.
# Fit model to each simulated dataset
simulation_results <- simulated_data %>%
mutate(
results = map(data, fit_model) # Apply the model
) %>%
unnest(results) # Combine all results into a single table
## Take a look
simulation_results
#> # A tibble: 1,000 × 4
#> sim_id data beta0_hat beta1_hat
#> <int> <list> <dbl> <dbl>
#> 1 1 <tibble [100 × 2]> 0.897 1.95
#> 2 2 <tibble [100 × 2]> 0.970 1.95
#> 3 3 <tibble [100 × 2]> 0.937 2.20
#> 4 4 <tibble [100 × 2]> 1.11 2.00
#> 5 5 <tibble [100 × 2]> 0.983 1.98
#> 6 6 <tibble [100 × 2]> 0.979 2.02
#> 7 7 <tibble [100 × 2]> 0.940 1.99
#> 8 8 <tibble [100 × 2]> 1.04 1.96
#> 9 9 <tibble [100 × 2]> 1.03 1.98
#> 10 10 <tibble [100 × 2]> 1.08 1.94
#> # ℹ 990 more rows
#> # ℹ Use `print(n = ...)` to see more rowsSummarize the results
# Summarize the results
summary_stats <- simulation_results %>%
summarize(
beta0_mean = mean(beta0_hat),
beta1_mean = mean(beta1_hat),
beta0_bias = mean(beta0_hat - 1),
beta1_bias = mean(beta1_hat - 2),
beta0_sd = sd(beta0_hat),
beta1_sd = sd(beta1_hat)
)
summary_stats
#> # A tibble: 1 × 6
#> beta0_mean beta1_mean beta0_bias beta1_bias beta0_sd beta1_sd
#> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 1.00 2.00 0.00336 0.00357 0.102 0.103Visualize the results (optional)
# Histogram of beta1 estimates
ggplot(simulation_results, aes(x = beta1_hat)) +
geom_histogram(bins = 30, fill = "skyblue", color = "black") +
geom_vline(xintercept = 2, linetype = "dashed", color = "red") +
labs(
title = "Distribution of Beta1 Estimates",
x = "Beta1 Estimates",
y = "Frequency"
) +
theme_minimal()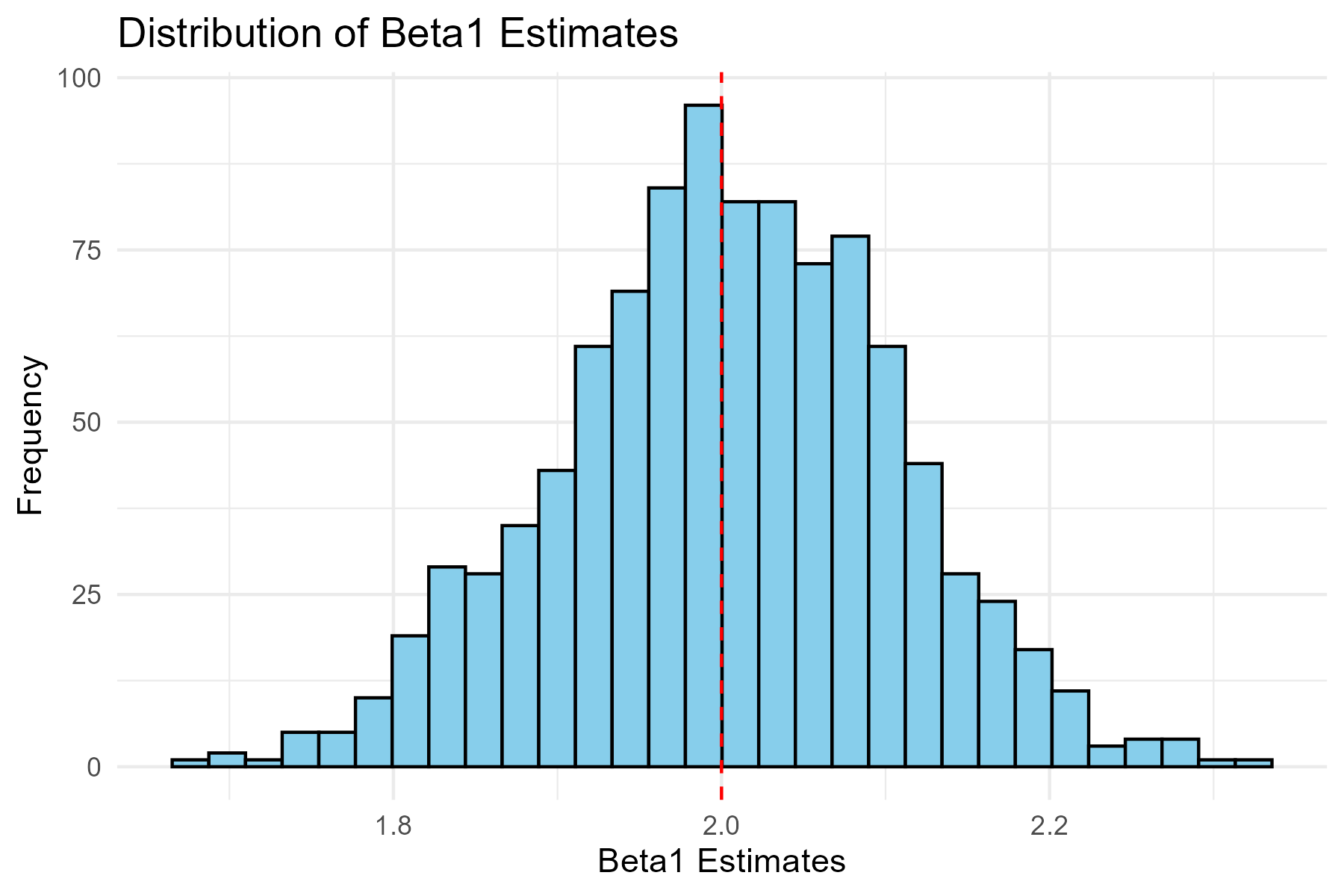
Key features of workflow
- Reproducibility:
- Use
set.seed()for consistent results.
- Use
- Scalability:
- Adjust
Nto increase or decrease the number of datasets.
- Adjust
- Summarization:
map()andunnest()make it easy to handle multiple datasets and their outputs.
- Customization:
- Replace the data-generating process or statistical method to fit your needs.