Tidy Survival Analysis: Applying R’s Tidyverse to Survival Data
Module 4. Semiparametric Regression Analysis
Lu Mao
Department of Biostatistics & Medical Informatics
University of Wisconsin-Madison
Aug 3, 2025
Presenting Regression Results
Cox PH Regression
- Model specification \[
\lambda(t \mid Z) = \lambda_0(t) \exp(\beta_1 Z_1 + \beta_2 Z_2 + \ldots + \beta_p Z_p)
\]
- \(\lambda_0(t)\): baseline hazard function
- \(\exp(\beta_j)\): hazard ratio for covariate \(Z_j\)
- GBC data: relapse-free survival
GBC Data: a Running Example
- Reformat the data
df <- gbc |> # calculate time to first event (relapse or death)
group_by(id) |> # group by id
arrange(time) |> # sort rows by time
slice(1) |> # get the first row within each id
ungroup() |> # remove grouping
mutate(
age40 = ifelse(age >= 40, 1, 0), # create binary variable for age >= 40
grade = factor(grade), # convert grade to factor
prog = prog / 100, # rescale progesterone receptor
estrg = estrg / 100 # rescale estrogen receptor
) Analysis in Base R
- Model fitting:
survival::coxph()
library(survival) # Load survival package
cox_fit <- coxph(Surv(time, status) ~ hormone + meno + age40 + grade + size + prog + estrg,
data = df)
summary(cox_fit) # Print model summaryCall:
coxph(formula = Surv(time, status) ~ hormone + meno + age40 +
grade + size + prog + estrg, data = df)
n= 686, number of events= 299
coef exp(coef) se(coef) z Pr(>|z|)
hormone -0.37432 0.68776 0.12917 -2.898 0.003758 **
meno 0.28450 1.32909 0.13973 2.036 0.041748 *
age40 -0.55127 0.57622 0.20243 -2.723 0.006463 **
grade2 0.71547 2.04514 0.24854 2.879 0.003993 **
grade3 0.77465 2.16982 0.26970 2.872 0.004075 **
size 0.01606 1.01619 0.00368 4.365 1.27e-05 ***
prog -0.22400 0.79932 0.05776 -3.878 0.000105 ***
estrg 0.01204 1.01212 0.04680 0.257 0.796895
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
exp(coef) exp(-coef) lower .95 upper .95
hormone 0.6878 1.4540 0.5339 0.8859
meno 1.3291 0.7524 1.0107 1.7478
age40 0.5762 1.7355 0.3875 0.8568
grade2 2.0451 0.4890 1.2565 3.3287
grade3 2.1698 0.4609 1.2790 3.6812
size 1.0162 0.9841 1.0089 1.0236
prog 0.7993 1.2511 0.7138 0.8951
estrg 1.0121 0.9880 0.9234 1.1093
Concordance= 0.661 (se = 0.016 )
Likelihood ratio test= 78.79 on 8 df, p=9e-14
Wald test = 68.82 on 8 df, p=8e-12
Score (logrank) test = 69.51 on 8 df, p=6e-12Tidy coxph() Output
- Using
broompackage:broom::tidy()- Provides a tidy data frame for easy manipulation and visualization
library(broom) # Load broom package
tidy_cox <- tidy(cox_fit) # Tidy the coxph output
tidy_cox # Display the tidy output# A tibble: 8 × 5
term estimate std.error statistic p.value
<chr> <dbl> <dbl> <dbl> <dbl>
1 hormone -0.374 0.129 -2.90 0.00376
2 meno 0.284 0.140 2.04 0.0417
3 age40 -0.551 0.202 -2.72 0.00646
4 grade2 0.715 0.249 2.88 0.00399
5 grade3 0.775 0.270 2.87 0.00408
6 size 0.0161 0.00368 4.37 0.0000127
7 prog -0.224 0.0578 -3.88 0.000105
8 estrg 0.0120 0.0468 0.257 0.797 Tabulating Results with gtsummary (I)
- Using
gtsummarypackage:tbl_regression()- Automatically formats regression results into a publication-ready table
library(gtsummary) # Load gtsummary package
cox_tbl <- cox_fit |> tbl_regression( # Create a regression table
exponentiate = TRUE, # Exponentiate coefficients to get hazard ratios
label = list(hormone ~ "Hormone Therapy", # Custom labels
meno ~ "Menopausal",
age40 ~ "Older than 40",
grade ~ "Tumor Grade",
size ~ "Tumor Size (mm)",
prog ~ "Progesterone Receptor (100 fmol/ml)",
estrg ~ "Estrogen Receptor (100 fmol/ml)")
) |>
add_global_p() # Add global p-value for categorical variables
cox_tbl # Display the regression tableTabulating Results with gtsummary (II)
- Result
| Characteristic | HR1 | 95% CI1 | p-value |
|---|---|---|---|
| Hormone Therapy | 0.69 | 0.53, 0.89 | 0.003 |
| Menopausal | 1.33 | 1.01, 1.75 | 0.039 |
| Older than 40 | 0.58 | 0.39, 0.86 | 0.009 |
| Tumor Grade | 0.004 | ||
| 1 | — | — | |
| 2 | 2.05 | 1.26, 3.33 | |
| 3 | 2.17 | 1.28, 3.68 | |
| Tumor Size (mm) | 1.02 | 1.01, 1.02 | <0.001 |
| Progesterone Receptor (100 fmol/ml) | 0.80 | 0.71, 0.90 | <0.001 |
| Estrogen Receptor (100 fmol/ml) | 1.01 | 0.92, 1.11 | 0.8 |
| 1 HR = Hazard Ratio, CI = Confidence Interval | |||
Further Customization
- Styling functions
modify_header(): update column headersmodify_footnote_header(): update column header footnotemodify_footnote_body(): update table body footnotemodify_caption(): update table caption/titlebold_labels(): bold variable labelsbold_levels(): bold variable levelsitalicize_labels(): italicize variable labelsitalicize_levels(): italicize variable levelsbold_p(): bold significant p-values
- More about
tbl_regression()
Table Customization Exercise
- Task: Customize the regression table
- Add a caption: “Cox regression analysis of the German breast cancer study”
- Bold significant p-values
- Italicize tumor grade levels
Other Regression Models
- Accelerated failure time (AFT) models \[
\log T = \beta_1 Z_1 + \beta_2 Z_2 + \ldots + \beta_p Z_p + \epsilon
\]
- \(\epsilon\sim\) Weibull, lognormal, etc. (parametric models)
- \(\exp(\beta_j)\): acceleration factor for covariate \(Z_j\)
- Model fitting:
survival::survreg()
Exercise
- Tidy up the
survregobjectaft_fitusingbroom::tidy() - Create a regression table using
gtsummary::tbl_regression()
Visualizing Hazard Ratios (I)
- Forest plot: Visualize hazard ratios and confidence intervals
# Tidy with exponentiated coeffs (HR) and CI
tidy_cox <- tidy(cox_fit, exponentiate = TRUE, conf.int = TRUE)
tidy_cox$term <- recode(tidy_cox$term, # Relabel the variables
hormone = "Hormone Therapy",
meno = "Menopausal",
age40 = "Older than 40",
grade2 = "Tumor Grade II vs I",
grade3 = "Tumor Grade III vs I",
size = "Tumor Size (mm)",
prog = "Progesterone (100 fmol/ml)",
estrg = "Estrogen (100 fmol/ml)")
tidy_cox |> # plot of hazard ratios and 95% CIs
ggplot(aes(y=term, x=estimate, xmin=conf.low, xmax=conf.high)) +
geom_pointrange() + # plots center point (x) and range (xmin, xmax)
geom_vline(xintercept=1, linetype = 2) + # vertical line at HR=1
scale_x_log10("Hazard ratio (95% CI)") + # log scale for x-axis
theme_classic() + # classic theme for clean look
theme(
axis.line.y = element_blank(), # remove y-axis line
axis.ticks.y = element_blank(), # remove y-axis ticks
axis.text.y = element_text(size = 11), # set variable label size
axis.title.y = element_blank() # remove y-axis title
)Visualizing Hazard Ratios (II)
- Result
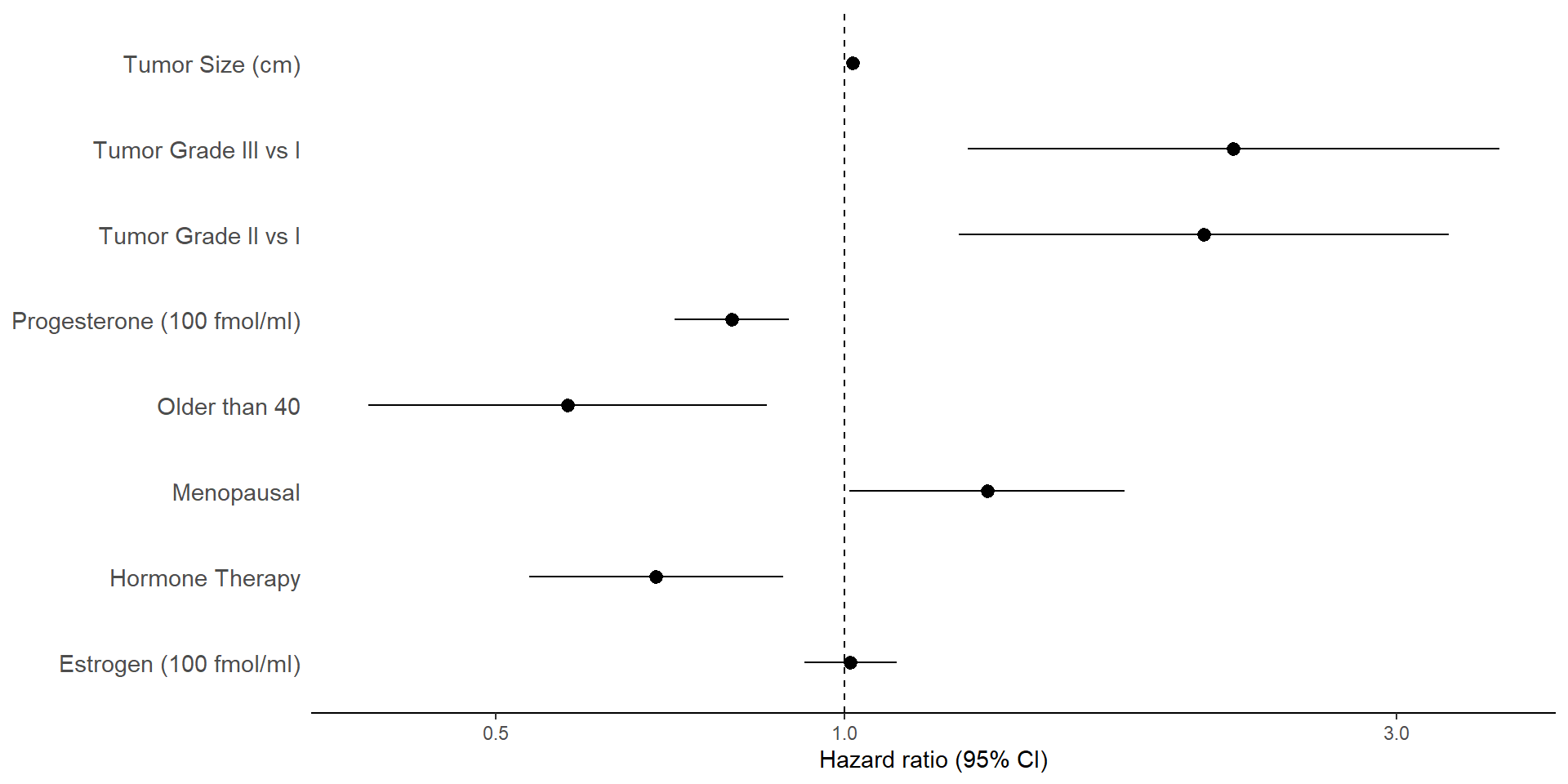
Forest Plot Exercise
- Task: customize the forest plot
- Use square rather than default circle for point estimates
- Set x-axis ticks at 0.5, 1, 2.0, and 4.0
- Add a title: “Cox Regression Results for GBC Data”
Solution
tidy_cox |>
ggplot(aes(y=term, x=estimate, xmin=conf.low, xmax=conf.high)) +
geom_pointrange(shape = 15) + # plots center point (x) in square and range (xmin, xmax)
geom_vline(xintercept=1, linetype = 2) + # vertical line at HR=1
scale_x_log10("Hazard ratio (95% CI)", # log scale for x-axis
breaks = c(0.5, 1, 2, 4)) + # log scale for x-axis
ggtitle("Cox Regression Results for GBC Data") + # Add title
theme_classic() + # classic theme for clean look
theme(
axis.line.y = element_blank(), # remove y-axis line
axis.ticks.y = element_blank(), # remove y-axis ticks
axis.text.y = element_text(size = 11), # set variable label size
axis.title.y = element_blank() # remove y-axis title
)Cox Model Prediction and Diagnostics
Model-Based Prediction
Predicted survival function \[ \hat S(t \mid z) = \exp\left\{- \exp(\hat\beta^\mathrm{T} z) \hat\Lambda_0(t)\right\} \]
Prepare new data for prediction
- A post-menpausal woman older than 40, undergoing hormone therapy, with tumor grade II, tumor size 20 mm, and progesterone and estrogen receptor levels both 100 fmol/ml.
Tidy Survival Prediction
Use
survival::survfit()to predict survival probabilitiesnewdata: new data for predictiontimes: time points for predictionbroom::tidy()to tidy the output
# Predict survival probabilities for `newdata` pred_surv <- survfit(cox_fit, newdata = new_data[1, ]) tidy_pred_surv <- tidy(pred_surv) # Tidy the survival prediction output head(tidy_pred_surv) # Display the first few rows of the tidy output# A tibble: 6 × 8 time n.risk n.event n.censor estimate std.error conf.high conf.low <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> 1 0.262 686 0 1 1 0 1 1 2 0.492 685 0 1 1 0 1 1 3 0.525 684 0 1 1 0 1 1 4 0.557 683 0 2 1 0 1 1 5 0.590 681 0 1 1 0 1 1 6 0.951 680 0 1 1 0 1 1
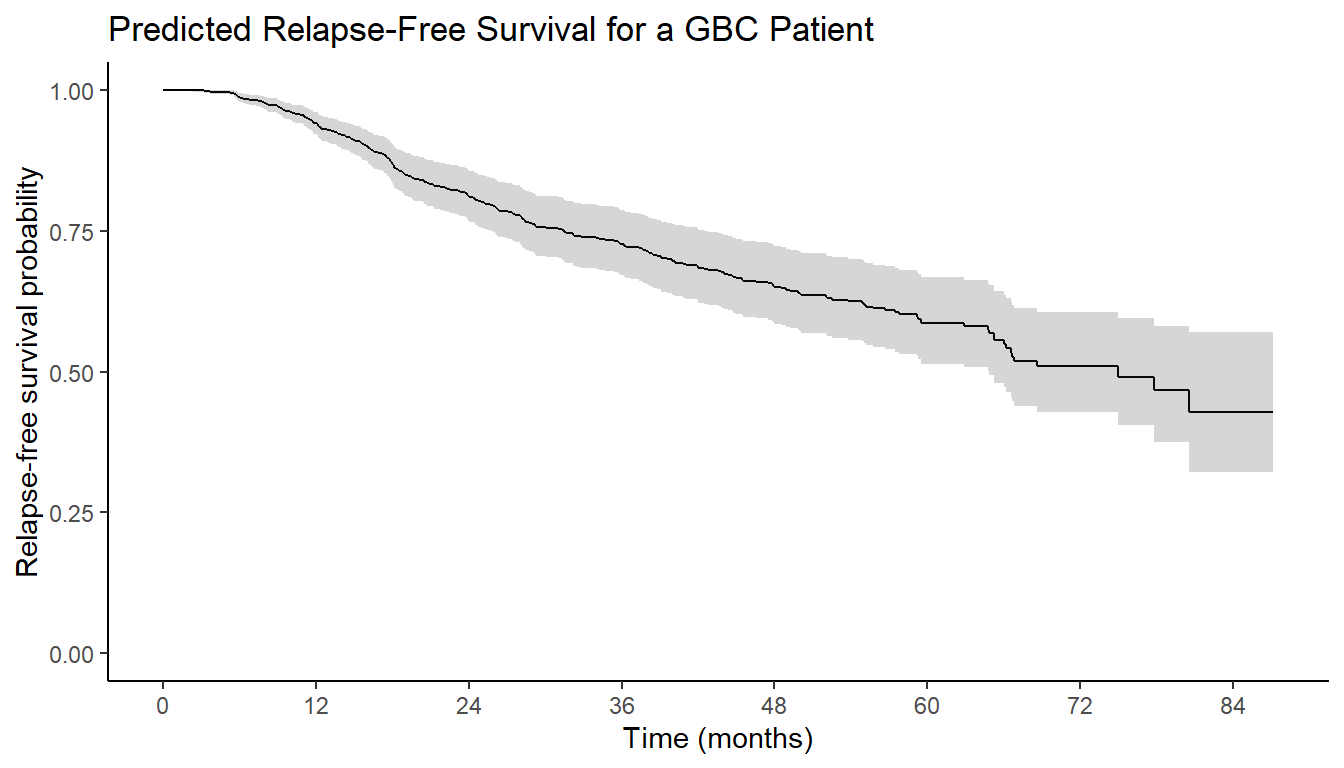
Visualizing Predicted Survival (I)
- Using
ggsurvfitpackage:ggsurvfit()- Pass
survfitobject toggsurvfit() - Similar customization to KM curves
- Pass
library(ggsurvfit) # Load ggsurvfit package
pred_fig <- pred_surv |> # Pass the survfit object
ggsurvfit() + # Main function
add_confidence_interval() + # Add confidence interval
scale_x_continuous("Time (months)", breaks = seq(0, 84, by = 12)) + # x-axis format
scale_y_continuous("Relapse-free survival probability", limits = c(0, 1)) + # y-axis format
ggtitle("Predicted Relapse-Free Survival for a GBC Patient") + # Add title
theme_classic() # Classic theme for clean lookVisualizing Predicted Survival (II)
- Result
Prediction Graphics Exercise
- Task: Add horizontal grid lines
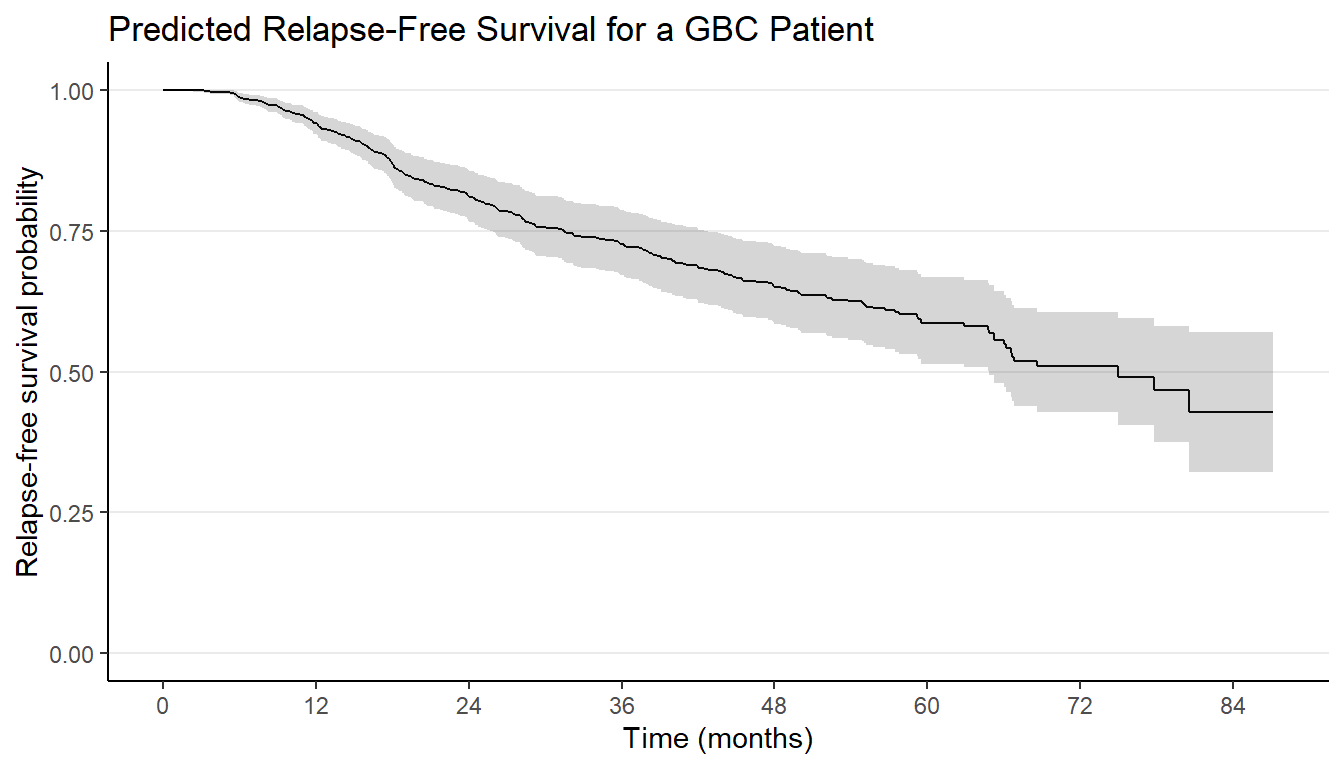
Cox Model Diagnostics
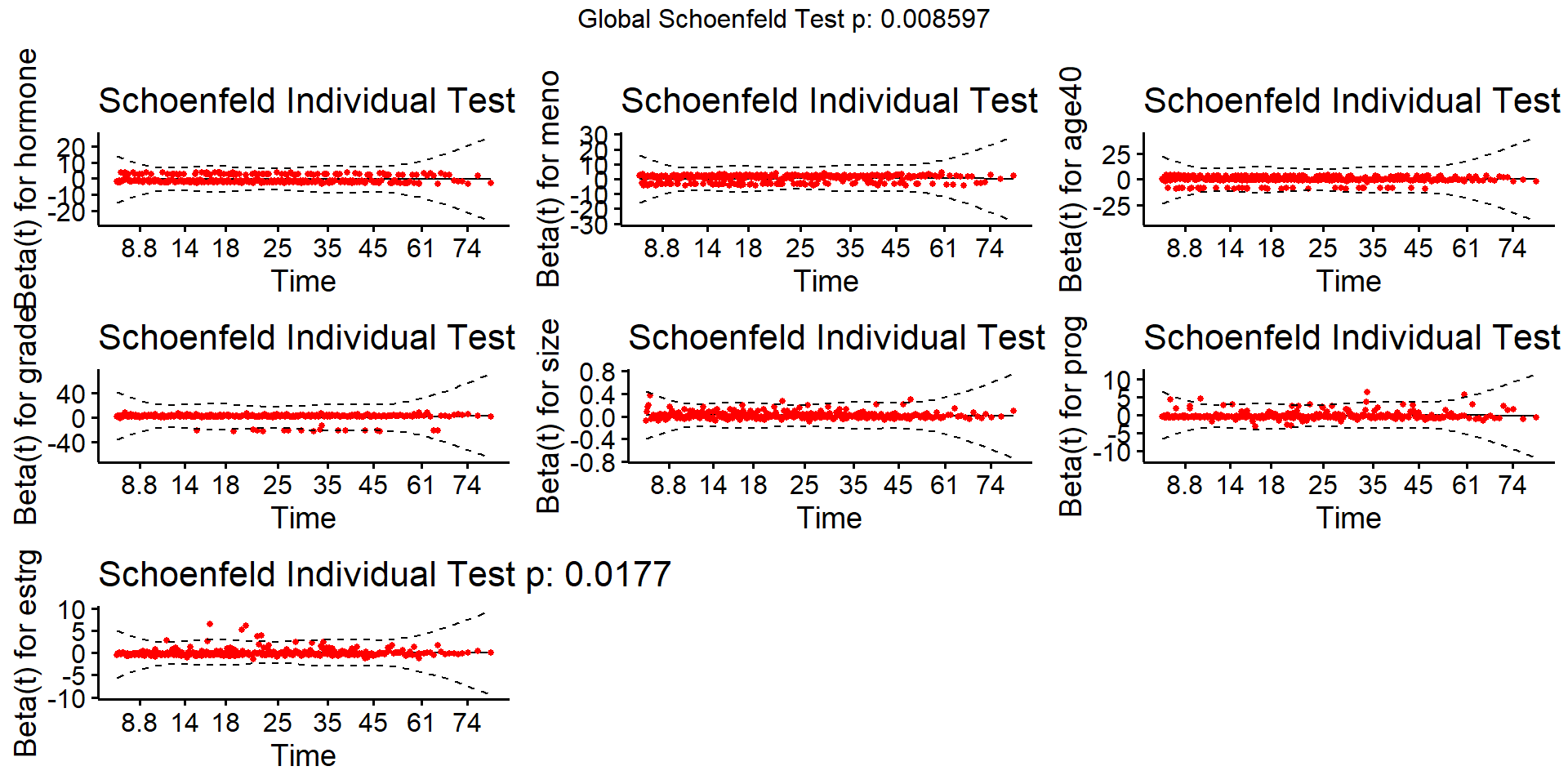
- PH assumptions: Schoenfeld residuals
- Difference between observed and expected covariate values at each event time
- Use
cox.zph()to test PH assumption - Use
survminer::ggcoxzph()oncox.zphobject to visualize Schoenfeld residuals
- Functional form of covariates
- Plot martingale residuals against (quantitative) covariates
- Use
residuals(cox_fit, type = "martingale")to get martingale residuals
- Other aspects
- Appropriateness of exponential link function
- Influential points/outliers
survminer::ggcoxdiagnostics()
Schoenfeld Residuals
Check proportionality
- Focus on graphics; use \(p\)-value only as guideline
Exponential Link Function
- Martingale vs. \(\hat\beta^\mathrm{T} Z_i\)
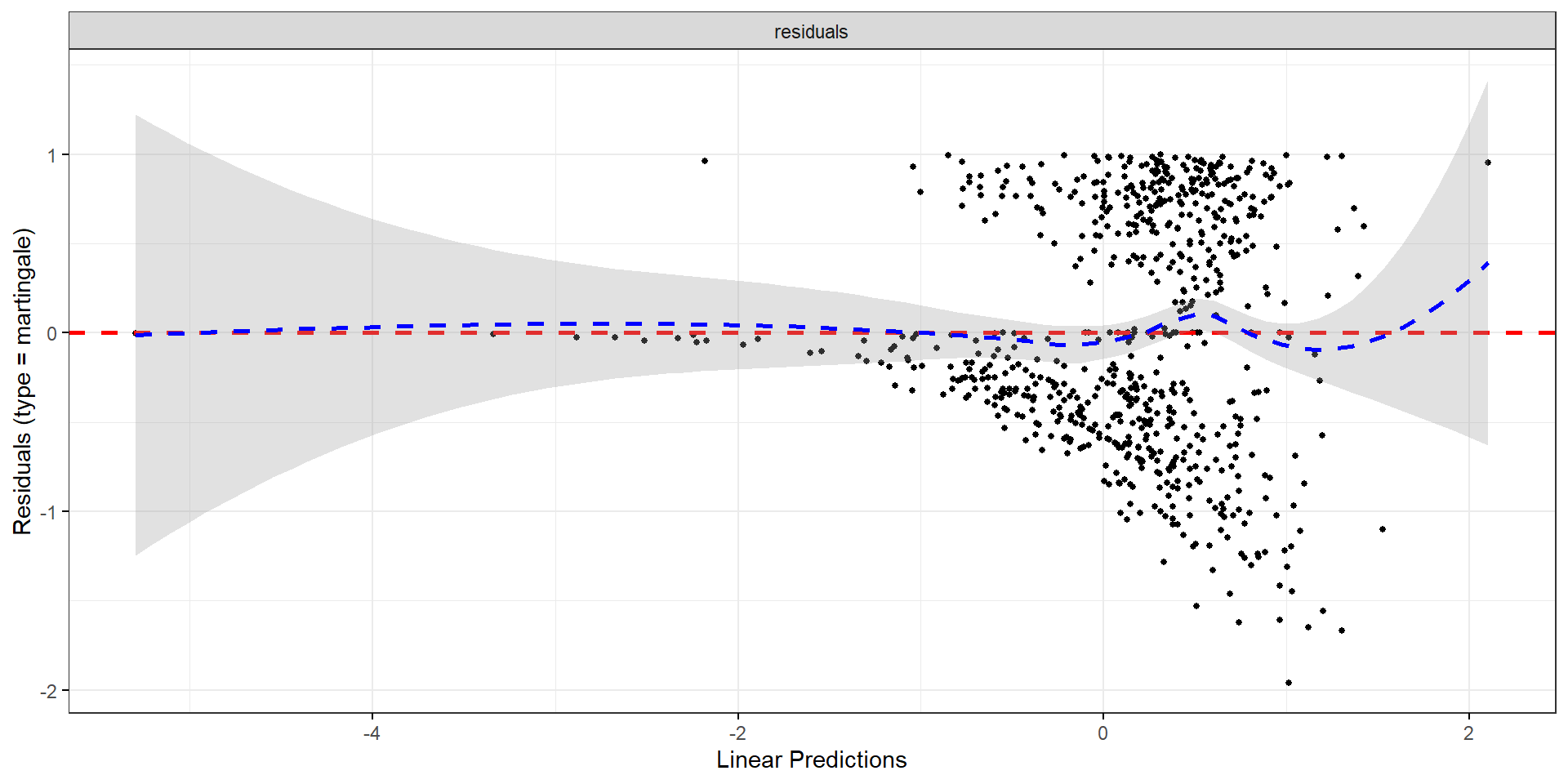
Influential Points
- Deviance residuals
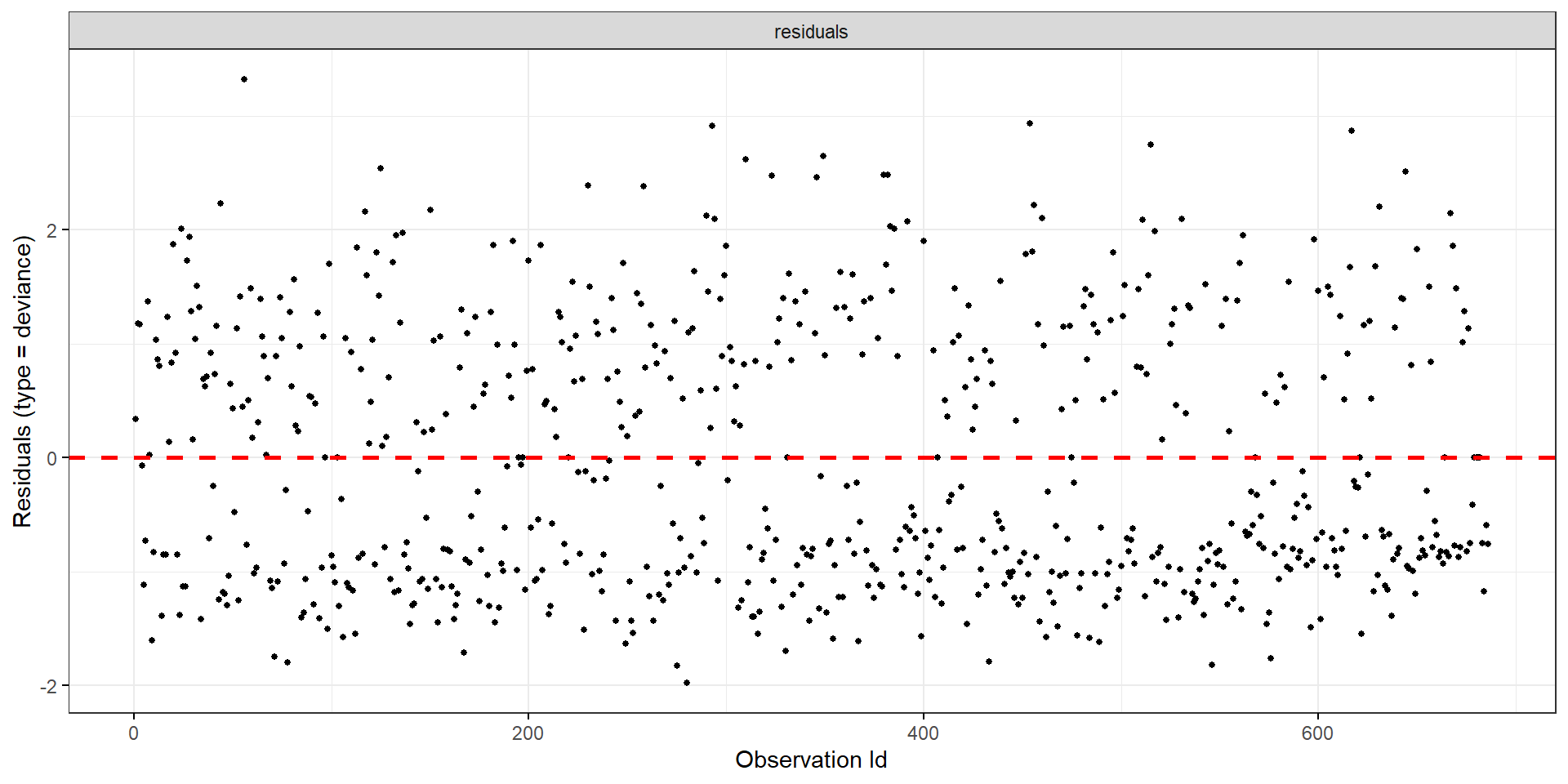
General Residual Graphics
- Basic arguments of
ggcoxdiagnostics()coxphobjecttype: Residual type (“martingale”, “deviance”, “score”, “schoenfeld”, “dfbeta”, “dfbetas”, and “scaledsch”)`ox.scale: Scale for x-axis (“linear.predictions”, “observation.id”, “time”)point.col: Color of pointspoint.size: Size of points- etc.
- More about
survminer- survminer website
Competing Risks Regression
Sub-Distribution Hazard
- Definition \[
\Lambda_k(t\mid Z) = -\log\left\{1 - F_k(t\mid Z)\right\}
\]
- \(F_k(t \mid Z)\): cumulative incidence function (CIF) of the \(k\)-th cause
- \(\lambda_k(t \mid Z)=\Lambda_k'(t\mid Z)\): risk of the \(k\)-th cause in presence of other competing events in the whole population
- Different from cause-specific hazard
- Cause-specific hazard \[\lambda^\mathrm{c}_k(t \mid Z) = \Pr(t\leq T< t+ \mathrm{d}t, \Delta = k\mid T\geq t, Z)/\mathrm{d}t \]
- Risk of the \(k\)-th cause in survivors
Fine-Gray Model
- Proportional sub-distribution hazards \[
\lambda_k(t \mid Z) = \lambda_0(t) \exp(\beta_1 Z_1 + \beta_2 Z_2 + \ldots + \beta_p Z_p)\]
- \(\lambda_0(t)\): baseline sub-distribution hazard function
- \(\exp(\beta_j)\): sub-distribution hazard ratio for covariate \(Z_j\)
library(tidycmprsk) # Load tidycmprsk package
data("trial", package = "tidycmprsk") # Load trial data from tidycmprsk package
head(trial) # Display the first few rows of the data# A tibble: 6 × 9
trt age marker stage grade response death death_cr ttdeath
<chr> <dbl> <dbl> <fct> <fct> <int> <int> <fct> <dbl>
1 Drug A 23 0.16 T1 II 0 0 censor 24
2 Drug B 9 1.11 T2 I 1 0 censor 24
3 Drug A 31 0.277 T1 II 0 0 censor 24
4 Drug A NA 2.07 T3 III 1 1 death other causes 17.6
5 Drug A 51 2.77 T4 III 1 1 death other causes 16.4
6 Drug B 39 0.613 T4 I 0 1 death from cancer 15.6Fitting Fine-Gray Model
- Using
cmprsk::crr()formula:Surv(time, status) ~ covariatesstatus: a factor with first level indicating censoring and subsequent levels the competing risks
failcode: event code for the cause of interest
fg_fit <- crr(Surv(ttdeath, death_cr) ~ trt + age + marker + stage, # fit FG model
failcode = "death from cancer", trial) # for death from cancer21 cases omitted due to missing values
Variable Coef SE HR 95% CI p-value
trtDrug B 0.396 0.283 1.49 0.85, 2.59 0.16
age 0.009 0.011 1.01 0.99, 1.03 0.42
marker -0.002 0.159 1.00 0.73, 1.36 0.99
stageT2 0.140 0.475 1.15 0.45, 2.92 0.77
stageT3 0.500 0.460 1.65 0.67, 4.06 0.28
stageT4 0.959 0.418 2.61 1.15, 5.91 0.022 Parameter Estimates and Variance
- Extracting \(\hat\beta\) and \(\hat{\mathrm{var}}(\hat\beta)\)
trtDrug B age marker stageT2 stageT3 stageT4
0.396486121 0.008956935 -0.002006717 0.140251967 0.500394141 0.958640247 [,1] [,2] [,3] [,4] [,5]
[1,] 0.0800665101 0.0001535045 -0.0051801922 0.011605373 0.0094601803
[2,] 0.0001535045 0.0001239790 -0.0005094795 0.001111245 -0.0009368412
[3,] -0.0051801922 -0.0005094795 0.0251827414 -0.028697647 -0.0037297926
[4,] 0.0116053732 0.0011112447 -0.0286976466 0.225187822 0.1101888313
[5,] 0.0094601803 -0.0009368412 -0.0037297926 0.110188831 0.2111942725
[6,] 0.0219362509 0.0010459739 -0.0176113331 0.124589145 0.1064088264
[,6]
[1,] 0.021936251
[2,] 0.001045974
[3,] -0.017611333
[4,] 0.124589145
[5,] 0.106408826
[6,] 0.174446817Tidy Fine-Gray Model Output
- Using
broompackage:broom::tidy()- Provides a tidy data frame for easy manipulation and visualization
tidy_fg <- tidy(fg_fit, exponentiate = TRUE, conf.int = TRUE) # Tidy model output
tidy_fg # Display the tidy output# A tibble: 6 × 7
term estimate std.error statistic conf.low conf.high p.value
<chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 trtDrug B 1.49 0.283 1.40 0.854 2.59 0.16
2 age 1.01 0.0111 0.804 0.987 1.03 0.42
3 marker 0.998 0.159 -0.0126 0.731 1.36 0.99
4 stageT2 1.15 0.475 0.296 0.454 2.92 0.77
5 stageT3 1.65 0.460 1.09 0.670 4.06 0.28
6 stageT4 2.61 0.418 2.30 1.15 5.91 0.022Forest Plot Exercise
- Task: Visualize sub-distribution hazard ratios and confidence intervals
Solution
# tidy_fg$term <- recode(tidy_fg$term, ...) # Relabel the variables
tidy_fg |> # plot of sub-distribution hazard ratios and 95% CIs
ggplot(aes(y=term, x=estimate, xmin=conf.low, xmax=conf.high)) +
geom_pointrange() + # plots center point (x) and range (xmin, xmax)
geom_vline(xintercept=1, linetype = 2) + # vertical line at HR=1
scale_x_log10("Sub-distribution hazard ratio (95% CI)") + # log scale for x-axis
theme_classic() + # classic theme for clean look
theme(
axis.line.y = element_blank(), # remove y-axis line
axis.ticks.y = element_blank(), # remove y-axis ticks
axis.text.y = element_text(size = 11), # set variable label size
axis.title.y = element_blank() # remove y-axis title
)FG Regression Table (I)
- Using
gtsummarypackage:tbl_regression()- Similarly to tabulating fitted
coxphobject
- Similarly to tabulating fitted
FG Regression Table (II)
- Result
| Characteristic | HR1 | 95% CI1 | p-value |
|---|---|---|---|
| Chemotherapy Treatment | 0.2 | ||
| Drug A | — | — | |
| Drug B | 1.49 | 0.85, 2.59 | |
| Age | 1.01 | 0.99, 1.03 | 0.4 |
| Marker Level (ng/mL) | 1.00 | 0.73, 1.36 | >0.9 |
| T Stage | 0.058 | ||
| T1 | — | — | |
| T2 | 1.15 | 0.45, 2.92 | |
| T3 | 1.65 | 0.67, 4.06 | |
| T4 | 2.61 | 1.15, 5.91 | |
| 1 HR = Hazard Ratio, CI = Confidence Interval | |||
Model-Based Prediction
- Predicted cumulative incidence function (CIF) \[ \hat F_k(t \mid z) = 1 - \exp\left\{-\hat\Lambda_k(t \mid z)\right\} \]
# Predict cumulative incidence function for first 10 patients
fg_pred <- predict(fg_fit, newdata= trial[1:10, ], times = c(6, 12, 18)) # Predict CIF
fg_pred # Display the predicted CIF$`time 6`
[1] 0.002279375 0.003430702 0.002447917 NA 0.007579470 0.010150002
[7] 0.002582498 0.002462688 0.002448570 0.006155050
$`time 12`
[1] 0.02093164 0.03135502 0.02246374 NA 0.06809922 0.09023665
[7] 0.02368558 0.02259790 0.02246967 0.05562630
$`time 18`
[1] 0.07090010 0.10483563 0.07594466 NA 0.21744137 0.28018810
[7] 0.07995368 0.07638548 0.07596414 0.18042196Summary
Key Takeaways
- Cox proportional hazards regression
- Tidy output with
broomandgtsummary - Visualize hazard ratios with forest plots with
ggplot2 - Model-based prediction with
survival::survfit()andggsurvfit() - Model diagnostics with
survminer
- Tidy output with
- Fine-Gray model for competing risks regression
- Fit with
cmprsk::crr() - Tidy output with
broomandgtsummary - Visualize sub-distribution hazard ratios with forest plots
- Fit with
Next Steps
- Machine learning: build best predictive model with many predictors
- Regularized Cox regression
- Parametric AFT models
- Survival trees
tidymodelspackages (censored)