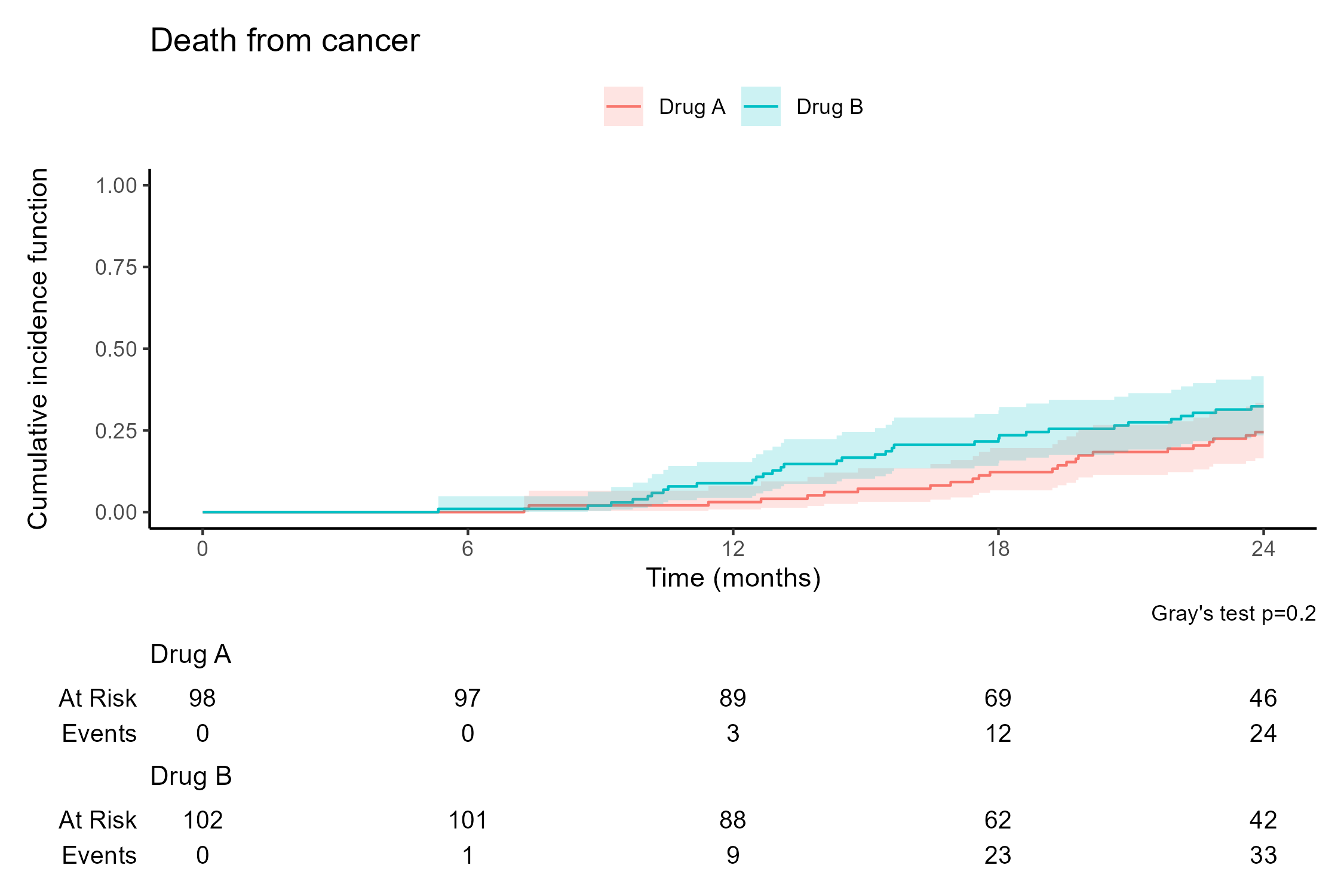
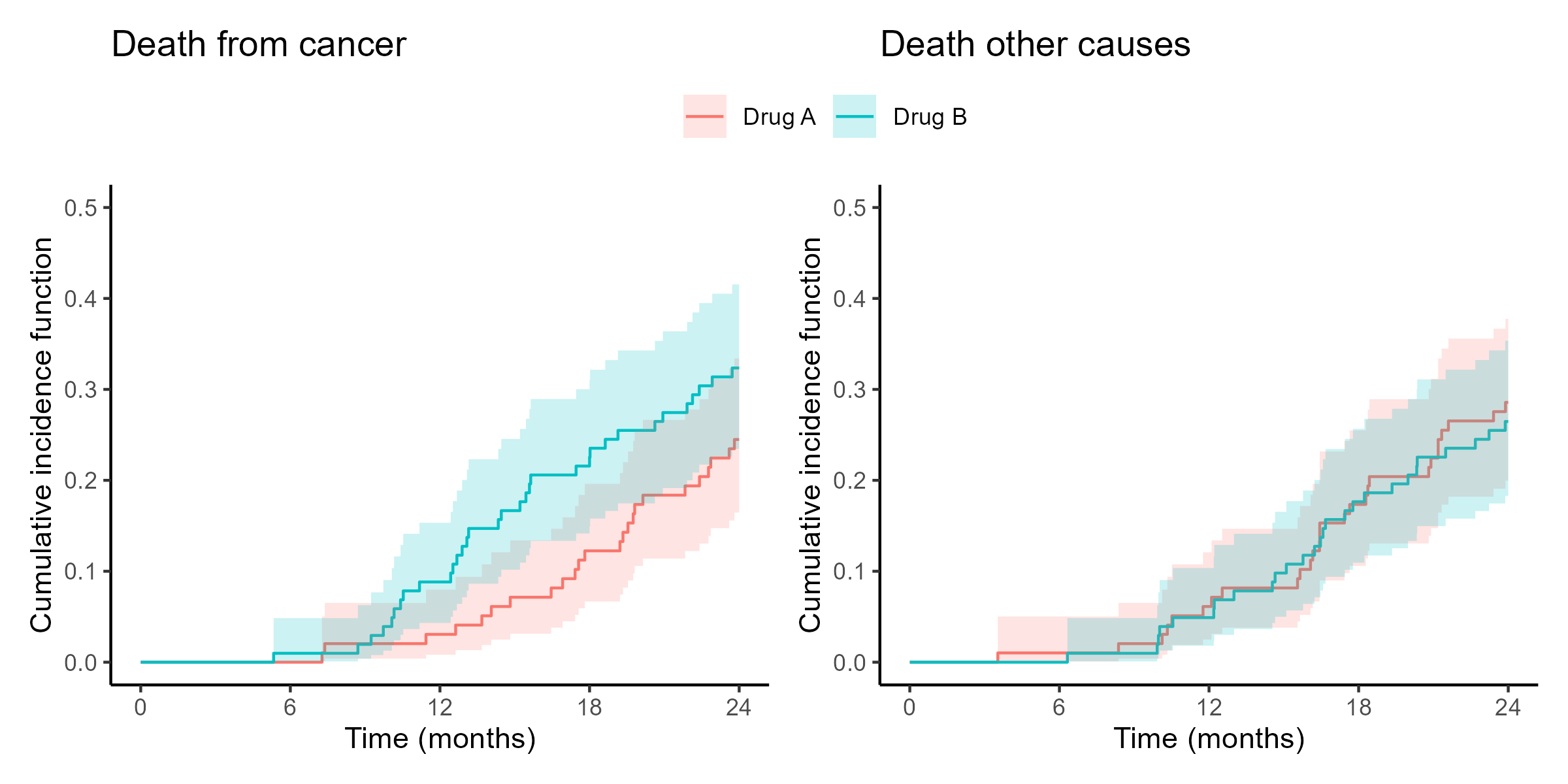
library(tidyverse) # Load tidyverse packages
# Load mortality + relapse data
gbc <- read.table("data/gbc.txt", header = TRUE)
df <- gbc |> # calculate time to first event (relapse or death)
group_by(id) |> # group by id
arrange(time) |> # sort rows by time
slice(1) |> # get the first row within each id
ungroup() # remove grouping
# Display the first few rows of the data
head(df) # A tibble: 6 × 11
id time status hormone age meno size grade nodes prog estrg
<int> <dbl> <int> <int> <int> <int> <int> <int> <int> <int> <int>
1 1 43.8 1 1 38 1 18 3 5 141 105
2 2 46.6 1 1 52 1 20 1 1 78 14
3 3 41.9 1 1 47 1 30 2 1 422 89
4 4 4.85 0 1 40 1 24 1 3 25 11
5 5 61.1 0 2 64 2 19 2 1 19 9
6 6 63.4 0 2 49 2 56 1 3 356 64