Applied Survival Analysis
Chapter 6 - Sample Size Calculation and Study Design
Lu Mao
Department of Biostatistics & Medical Informatics
University of Wisconsin-Madison
Outline
Sample size for Cox model and RMST
Impact of study design and censoring
An example using pilot study
\[\newcommand{\d}{{\rm d}}\] \[\newcommand{\T}{{\rm T}}\] \[\newcommand{\dd}{{\rm d}}\] \[\newcommand{\pr}{{\rm pr}}\] \[\newcommand{\var}{{\rm var}}\] \[\newcommand{\se}{{\rm se}}\] \[\newcommand{\indep}{\perp \!\!\! \perp}\] \[\newcommand{\Pn}{n^{-1}\sum_{i=1}^n}\]
Sample Size Estimation
Motivation
- Sample size calculation
- Ensure power (80% or 90%) to detect meaningful difference
- Power: probability of rejecting \(H_0\) under alternative
- Too small sample \(\to\) low power \(\to\) fail to reject \(H_0\) even if \(H_A\) is true
- Ensure power (80% or 90%) to detect meaningful difference
- Input information
- Hypothesized effect size: minimal important difference (MID)
- Hazard ratio, difference in RMST, etc.
- Desired power (0.8 or 0.9)
- Significance level \(\alpha\) (0.05)
- Group allocation ratio (1:1)
- Hypothesized effect size: minimal important difference (MID)
General Setting
- Standardized test statistic \[\begin{equation}\label{eq:design_form}
S_n=\sqrt n T_n/\hat\sigma_n \stackrel{H_0}{\sim}\mathcal N(0, 1)
\end{equation}\]
- \(\sqrt n T_n\stackrel{H_0}{\sim}\mathcal N(0,\sigma_0^2)\); \(\,\) \(\hat\sigma^2_n=\) Estimator of \(\sigma_0^2\)
- \(T_n\): Test statistic converging at \(n^{-1/2}\) rate as \(n\to\infty\)
- Level-\(\alpha\) test
- Reject \(H_0\) if \(|S_n|>z_{1-\alpha/2}\)
- \(z_{1-\alpha/2}\): \(100(1-\alpha/2)\) percentile of standard normal
General Derivation
- Effect size \(\theta\)
- \(H_0: \theta = 0\); \(\,\) \(H_A: \theta\neq 0\)
- \(\theta=\) log-hazard ratio under Cox model
- \(\theta =\) difference in RMST
- \(H_0: \theta = 0\); \(\,\) \(H_A: \theta\neq 0\)
- Suppose under \(H_A\)
- Mean of test statistic (\(\theta=\) MID) \[
E_\theta(T_n)\approx f(\theta)
\]
- \(f(\theta)\) is some function with \(f(0)=0\) (centered under \(H_0\))
- Then, \[S_n=\sqrt n T_n/\hat\sigma_n \stackrel{H_A}{\sim} \mathcal N\{\sqrt nf(\theta)/\sigma_0, 1\}\]
- \(f(\theta)/\sigma_0\): signal-to-noise ratio
- Mean of test statistic (\(\theta=\) MID) \[
E_\theta(T_n)\approx f(\theta)
\]
General Formula
- Power requirement \[
\pr_\theta(|S_n|>z_{1-\alpha/2})\geq\gamma
\]
- \(\gamma\): desired power (0.8 or 0.9)
- Use \(S_n \sim \mathcal N\{\sqrt nf(\theta)/\sigma_0, 1\}\) to solve for \(n\)
- Sample size formula \[\begin{equation}\label{eq:design_ss_gen}
n=\frac{\sigma_0^2(z_{1-\alpha/2}+z_\gamma)^2}{f(\theta)^2}
\end{equation}\]
- Find mean \(f(\theta)\) and variance \(\sigma_0^2\)
Case Study: Cox Model
- Partial-likelihood score/log-rank test
- Test statistic \(T_n = U_n(0)\)
- Equivalent to log-rank test under \(Z = 1, 0\)
- \(f(\theta)=E\{U_n(0)\}\approx\theta q(1-q)\psi\) and \(\sigma_0^2=q(1-q)\psi\)
- \(q=N_1/n\): proportion randomized to treatment
- \(\theta\): log-hazard ratio
- \(\psi=\int_0^\infty \pi(t)\lambda_0(t)\dd t=\pr(T\leq C)\): proportion of failure observed, where \(\pi(t)=\pr(X> t)\)
- Test statistic \(T_n = U_n(0)\)
- Sample size formula \[\begin{equation}\label{eq:cox_sample_size}
n= \frac{(z_{1-\alpha/2}+z_{\gamma})^2}{q(1-q)\psi\theta^2}
\end{equation}\]
- Expected number of events \(n\psi = (z_{1-\alpha/2}+z_{\gamma})^2/\{q(1-q)\theta^2\}\)
Case Study: RMST
- Difference in \(\tau\)-RMST
- Test statistic \(T_n = \hat\theta(\tau)=\hat\mu_1(\tau) - \hat\mu_0(\tau)\)
- \(f(\theta)=E\{\hat\theta(\tau)\}\approx\theta(\tau)\) and \(\sigma_0^2=\var\{\sqrt n\hat\theta(\tau)\}\approx q^{-1}(1-q)^{-1}\zeta(\tau)\)
- \(\zeta(\tau)=\int_0^\tau R_0(\tau;t)^2\pi(t)^{-1}\lambda_0(t)\dd t\)
- \(R_0(\tau;t)=\int_t^\tau S_0(t)\dd t\)
- \(S_0(t)\) and \(\lambda_0(t)\): survival/hazard functions in control
- Test statistic \(T_n = \hat\theta(\tau)=\hat\mu_1(\tau) - \hat\mu_0(\tau)\)
- Sample size formula \[\begin{equation}\label{eq:rmst_sample_size} n= \frac{\zeta(\tau)(z_{1-\alpha/2}+z_{\gamma})^2}{q(1-q)\theta(\tau)^2} \end{equation}\]
Impact of Study Design
Dependence on Censoring
- Two types of parameters
- The failure factor: \(S_0(t), \lambda_0(t)\), log-HR, \(\theta(\tau)\)
- Use domain knowledge (or pilot study data) to specify (or estimate)
- The censoring factor \[
\pi(t)=\pr(X>t)=S_0(t)G(t)
\]
- \(G(t)=\pr(C > t)\)
- Involved in \(\psi\) for log-rank (Cox) and \(\zeta(\tau)\) for RMST
- The failure factor: \(S_0(t), \lambda_0(t)\), log-HR, \(\theta(\tau)\)
- Censoring depends on study design
- Initial period of subject accrual
- Additional follow-up
Simplified Model
- Parametric set-up
- \(T\sim\mbox{Expn}(\lambda_0)\)
- Loss to follow-up (LTFU) \(\sim\mbox{Expn}(\lambda_L)\)
- Administrative censoring \(\sim\mbox{Unif}[c, b + c]\)
- Uniform accrual in \([0, b]\)
- Subsequent follow-up time \(c\)
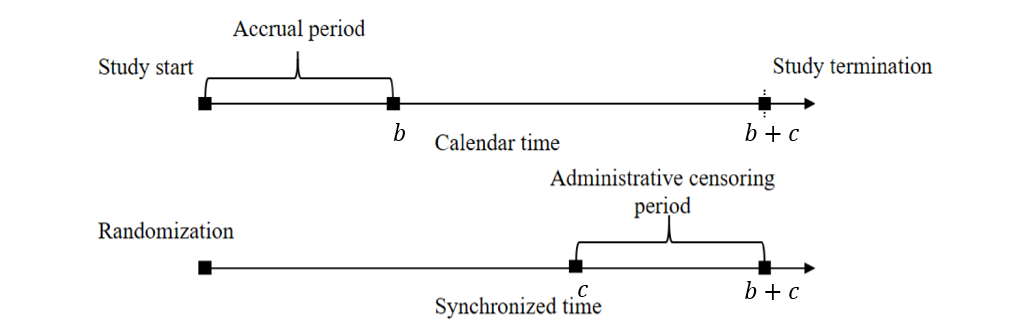
Model-Based Expressions
- Under this set-up
- Censoring survival function \[\begin{equation}\label{eq:design:censoring}
G(t;\lambda_L, b, c):=\pr(C>t)=\left\{\begin{array}{ll} \exp(-\lambda_L t)& 0\leq t\leq c\\
b^{-1}(c+b-t)\exp(-\lambda_L t) & c<t<c+b \\
0& t\geq c+b.
\end{array}\right.
\end{equation}\]
- \(\psi\) for log-rank (closed-form) \[\begin{equation}\label{eq:cox:psi} \psi(\lambda_0,\lambda_L, b, c)=\frac{\lambda_0}{\lambda_0+\lambda_L}\left[1-\exp\{-(\lambda_0+\lambda_L)c\} \frac{1-\exp\{-(\lambda_0+\lambda_L)b\}}{(\lambda_0+\lambda_L)b}\right] \end{equation}\]
- \(\zeta(\tau)\) for RMST (needs numerical integration) \[\begin{equation}\label{eq:design:rmst_zeta} \zeta(\tau; \lambda_0, \lambda_L, b, c)=\lambda_0^{-1}\int_0^\tau\{\exp(-\lambda_0 t)-\exp(-\lambda_0 \tau)\}^2\exp(\lambda_0 t)G(t;\lambda_L, b, c)^{-1}\dd t \end{equation}\]
- Plug \(\psi\) or \(\zeta(\tau)\) back into sample size formulas
- Censoring survival function \[\begin{equation}\label{eq:design:censoring}
G(t;\lambda_L, b, c):=\pr(C>t)=\left\{\begin{array}{ll} \exp(-\lambda_L t)& 0\leq t\leq c\\
b^{-1}(c+b-t)\exp(-\lambda_L t) & c<t<c+b \\
0& t\geq c+b.
\end{array}\right.
\end{equation}\]
Programming the Functions (I)
- \(G(t;\lambda_L, b, c)\): Survival function for censoring
- \(\psi(\lambda_0,\lambda_L, b, c)\): \(\psi\) parameter for log-rank
Programming the Functions (II)
- \(\zeta(\tau; \lambda_0, \lambda_L, b, c)\): \(\zeta(\tau)\) for RMST
# ------ integrand function --------------------------------
zeta_integrand <- function(t, tau, lambda0,lambdaL, b, c){
integrand <- (exp(- lambda0 * t) - exp( - lambda0 * tau))^2*
exp(lambda0 * t)/(Gfun(t, lambdaL, b, c) * lambda0)
return(integrand)
}
# ------ numerical integration by integrate() --------------
zeta_fun <- function(tau, lambda0, lambdaL, b, c){
f <- function(t){
return(zeta_integrand(t, tau, lambda0, lambdaL, b, c))
}
zeta <- integrate(f, lower = 0, upper = tau)
return(zeta$value)
}How to Specify Parameters
- Specification of \((\lambda_0, \lambda_L, b, c)\)
- \(\lambda_0\) (and \(\lambda_L\)): use domain knowledge or estimate from historical data
- \(\hat\lambda_0\): (crude) event rate (see Chapters 1 & 2) \[ \hat\lambda_0 = \frac{\mbox{Number of events}}{\mbox{Total person-time}} \]
- \(b, c\): determined by current study design
- Specification of \(\theta\)
- Supplied by investigator based on domain knowledge (MID)
An Example
Example: GBC as Pilot Study (I)
- Design objective
- A randomized controlled trial to evaluate hormonal treatment on relapse-free survival of post-menopausal breast cancer patients
- \(b=2\) years of accrual; \(c=3.5\) years subsequent follow-up
- GBC pilot data
- \(\hat\lambda_0=0.174\) per person-year based on \(n=209\)
- Other parameters
- Set \(\lambda_L=0.01\) per person-year
- \(\lambda_1\): (constant) hazard rate in treatment, taking a range of values
- HR: \(\lambda_1/\lambda_0\); difference in RMST (Exercise)
Example: GBC as Pilot Study (II)
- Sample sizes for Cox (log-rank), 3y-RMST, 5y-RMST
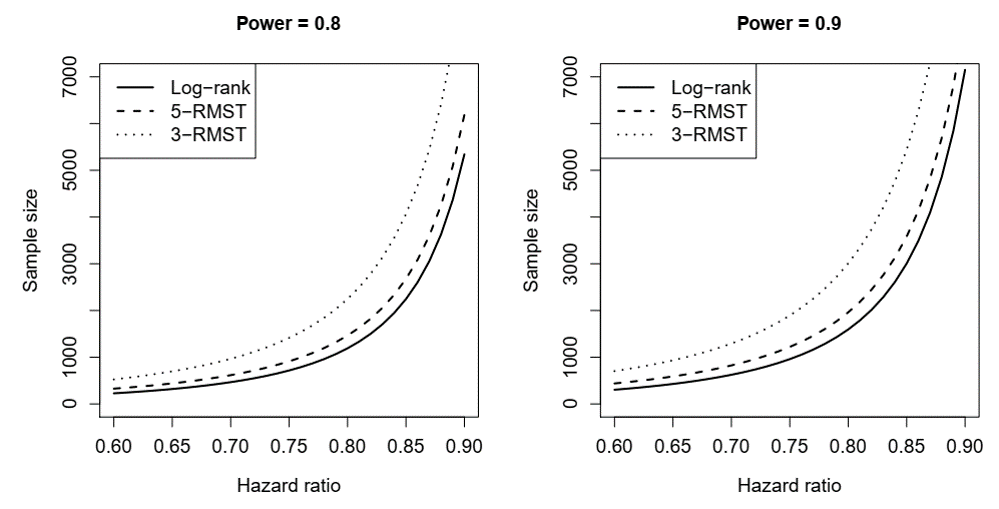
Conclusion
Notes (I)
- SAS code for log-rank
- Sample size formula for log-rank (Schoenfeld, 1981)
- Freedman (1982): more accurate when \(\theta\) is large
- Schoenfeld (1983): covariate adjusted test (Schoenfeld, 1983)
Notes (II)
R-package
npsurvSS(Yung and Liu, 2020)- Various tests (e.g., weighted log-rank, RMST, survival probability tests)
- Non-exponential event/ LTFU, non-uniform accrual
# Set up the two groups control <- create_arm( # Parameter set-up for control ) treatment <- create_arm( # Parameter set-up for treatment ) # Compute sample size size_two_arm(control, treatment, test = list(list(test="weighted logrank"), # Log-rank test list(test="rmst difference", milestone=3), # 3y-RMST list(test="rmst difference", milestone=5)) # 5y-RMST )
Summary
- Sample size calculations
- Cox (log-rank) \[\begin{equation} n= \frac{(z_{1-\alpha/2}+z_{\gamma})^2}{q(1-q)\psi\theta^2} \end{equation}\]
- \(\tau\)-RMST \[\begin{equation} n= \frac{\zeta(\tau)(z_{1-\alpha/2}+z_{\gamma})^2}{q(1-q)\theta(\tau)^2} \end{equation}\]
- \(\psi\) or \(\zeta(\tau)\) calculated under simplifying assumptions
- Exponential failure and LTFU
- Uniform accrual